Single-cell
Transcriptomics and beyond
The Single-cell Analysis team of SaeysLab develops open-source algorithms that leverage single-cell transcriptomics data to further our biological understanding of how cells cooperate across space and time to perform various functions.
How can prior biological knowledge and various omics technologies be combined to infer cell-cell communication processes, both within and across samples of healthy and diseased tissue?
To further advance the field of single-cell transcriptomics, this team also performs benchmarks of various algorithms and pipelines. What are the most suitable methods, for example to infer a trajectory or perform spatial deconvolution?
Highlighted single-cell papers
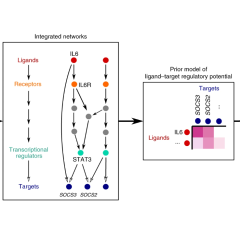
NicheNet: modeling intercellular communication by linking ligands to target genes
Robin Browaeys, Wouter Saelens & Yvan Saeys
Nature methods 2019
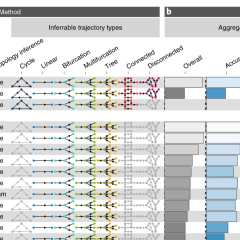
A comparison of single-cell trajectory inference methods
Wouter Saelens, Robrecht Cannoodt, Helena Todorov, Yvan Saeys
Nature Biotechnology 2019

